List of polymorphisms in rice core collections
[To the top page]
This page describes a list of polymorphisms among Japan and world core collection of rice distributed by NARO genebank. NGS data of core collection analyzed by PED program.
- To enlarge figure, click on the figure
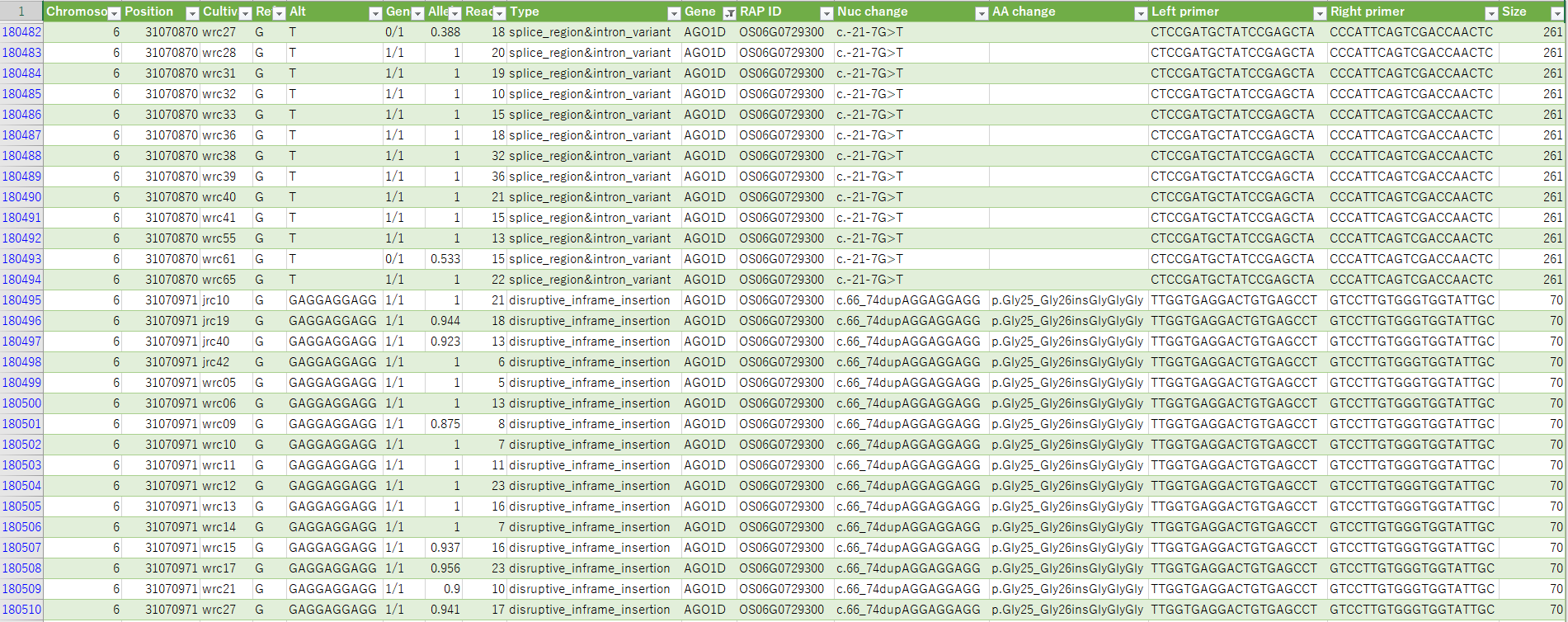
- Polymorphism lists in each chromosome will be downloaded from

- Nucleotide sequence of primer pairs amplified at the polymorphic loci also included in the list.
- Annotation of polymorphisms have been added with analysis of snpEff.
- Seeds of core collection are able to obtain from NARO genebank. When fortunately found cultivars which have suitable mutation, the mutation will be introduced by crossing.
- Information of SNPs are already published in TASKE+. TASKE+'s SNPs were detected by BWA-mem and GATK. Comparing between the PED list and TASKE+ information will help for evaluation of both system.
Due to high threshold value of PED, number of detected SNPs by PED is less than TASKE+. Detection of structural variations by PED is better than TASKE+.
Data import into Excel
- Files are text format. Columns are separated by tab suitable to import by Excel.
- List are saved into separated files with chromosome and archived into single zip file, jrc_wrc.zip. The file can be obtained from Genome wide variations of rice detected by PED page.
- Open in the data tab on Excel, import the chromosome file from text or CSV format.
- When open the Power Query, set Column 6 to text format.
- Select records belongs the target gene or IRGSP-1.0 accession by filtering with Column J (Gene name) or Column K (RapID).
- Nonsense or frameshift mutations may disrupt the gene function.
- Missense mutations cause no-effect of reduce (or increase in the rare case) the gene activity.
- Primer pairs which amplify the target region are provided in each mutation and can be amplified the target region for validation of the mutation.
Because length of amplified region is relatively short, the amplified fragments can be analyzed by high resolution melting analysis.
Reference
- Tanaka, N., Shenton, M., Kawahara, Y. et al. (2020). Whole-Genome Sequencing of the NARO World Rice Core Collection (WRC) as the Basis for Diversity and Association Studies. Plant & cell physiology, 61(5), 922-932. https://doi.org/10.1093/pcp/pcaa019
- Tanaka, N., Shenton, M., Kawahara, Y. et al. (2021). Investigation of the Genetic Diversity of a Rice Core Collection of Japanese Landraces using Whole-Genome Sequencing. Plant & cell physiology, 61(12), 2087-2096. https://doi.org/10.1093/pcp/pcaa125
- Kumagai, M., Nishikawa, D., Kawahara, Y. et al. (2019). TASUKE+: a web-based platform for exploring genome-wide association studies results and large-scale resequencing data. DNA research : an international journal for rapid publication of reports on genes and genomes, 26(6), 445-452. https://doi.org/10.1093/dnares/dsz022
- Cingolani, P., Platts, A., Wang, L. L., Coon, M., et al. (2012). A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly, 6(2), 80-92. https://doi.org/10.4161/fly.19695
- Miyao, A., Kiyomiya, J.S., Iida, K. et al. (2019). Polymorphic edge detection (PED): two efficient methods of polymorphism detection from next-generation sequencing data. BMC Bioinformatics 20, 362. https://doi.org/10.1186/s12859-019-2955-6
- Miyao, A. (2024). Genome wide variations of rice detected by PED [Data set]. Zenodo. https://doi.org/10.5281/zenodo.11159847
- https://github.com/akiomiyao/ped/
Contact
Akio Miyao (miyao@affrc.go.jp), NARO

