- Installing software required by PED
$ sudo apt update
$ sudo apt upgrade
$ sudo apt install curl
$ sudo apt install git
$ git clone https://github.com/akiomiyao/ped.git
- To download sequence data, fastq-dump from NCBI is required.
Tool kit can be download from
https://trace.ncbi.nlm.nih.gov/Traces/sra/sra.cgi?view=software
Details of setup fastq-dump is described in
https://akiomiyao.github.io/ped/sratoolkit/index.html
- Download fastq files of the Platinum genome sequence (ERR194146 and ERR194147) from NCBI Sequence read archive site.
$ cd ped
$ git pull
$ perl download.pl accession=ERR194146
$ perl download.pl accession=ERR194147
Scripts are updated by git pull command.
Files of fastq format are downloaded into ERR194146/read and ERR194147/read.
Downloading is slow. It requires one or two days.
Sometimes error message will appear. The script will try to reconnect. Check increasing size of downloading files.
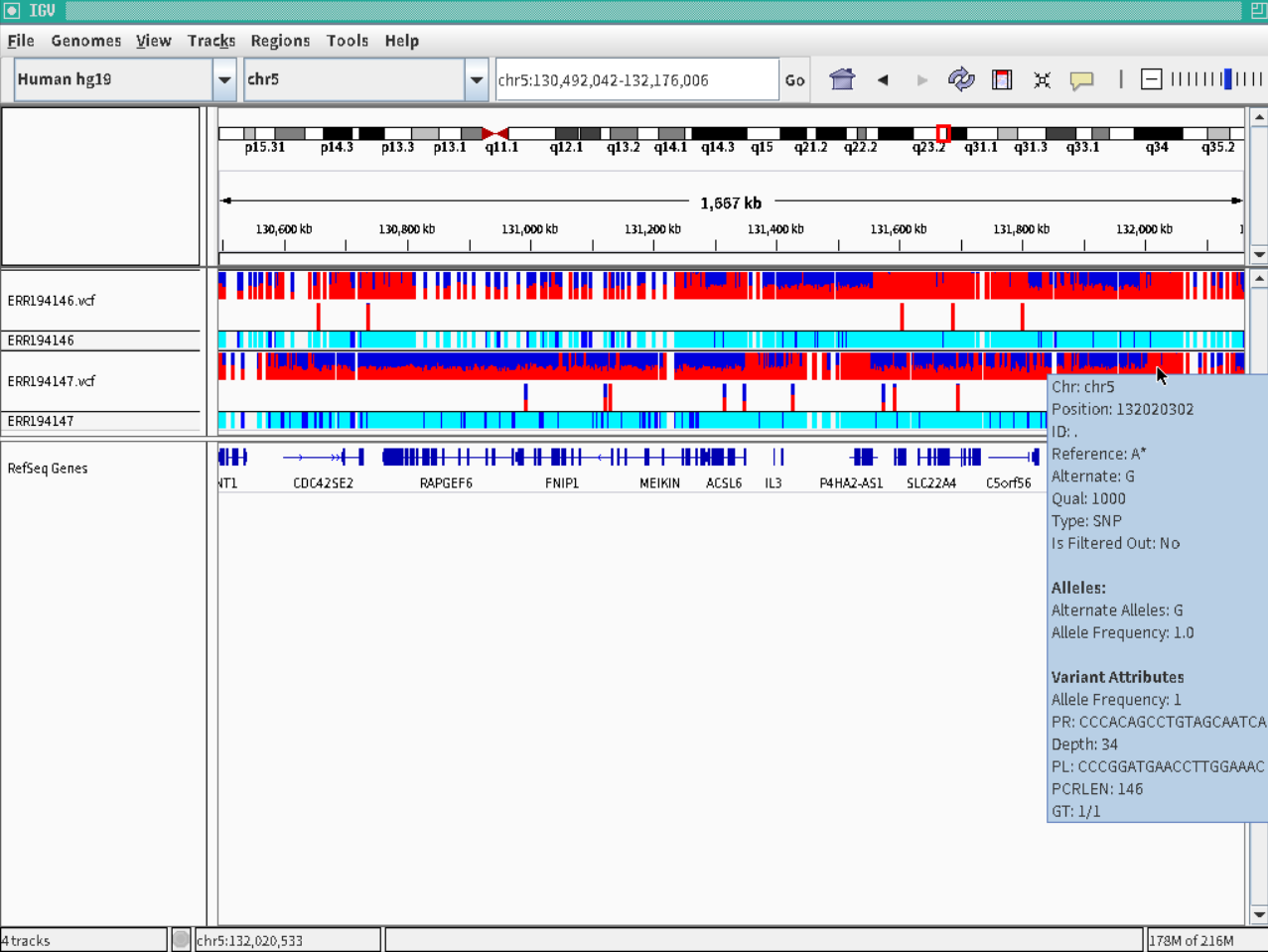
- Polymorphism detection of ERR194146 and ERR194147 sequences on human reference genome hg19.
$ perl ped.pl target=ERR194146,ref=hg19
$ perl ped.pl target=ERR194147,ref=hg19
Since hg19 reference file is already configured, data are automatically downloaded and calculated.
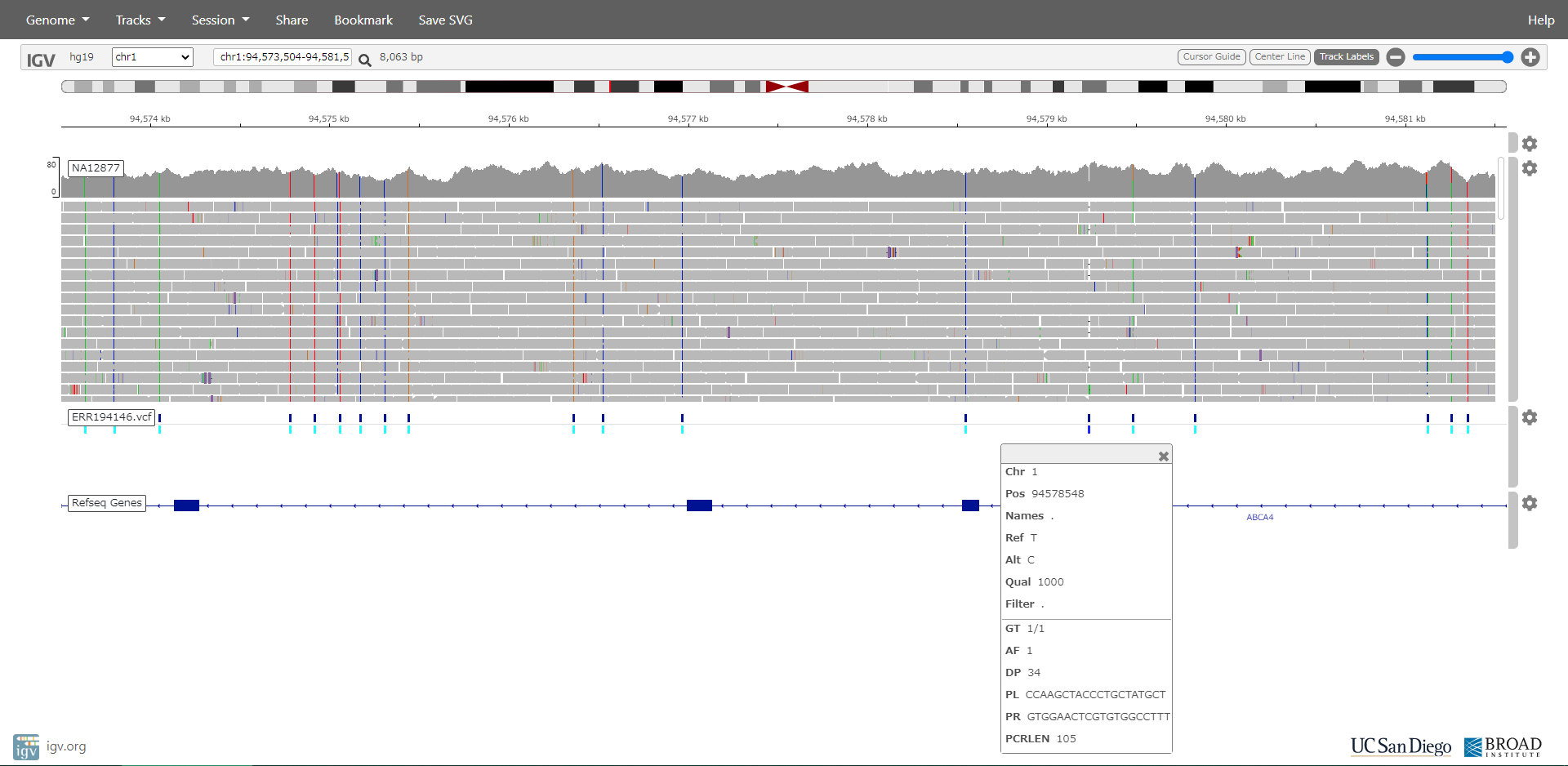
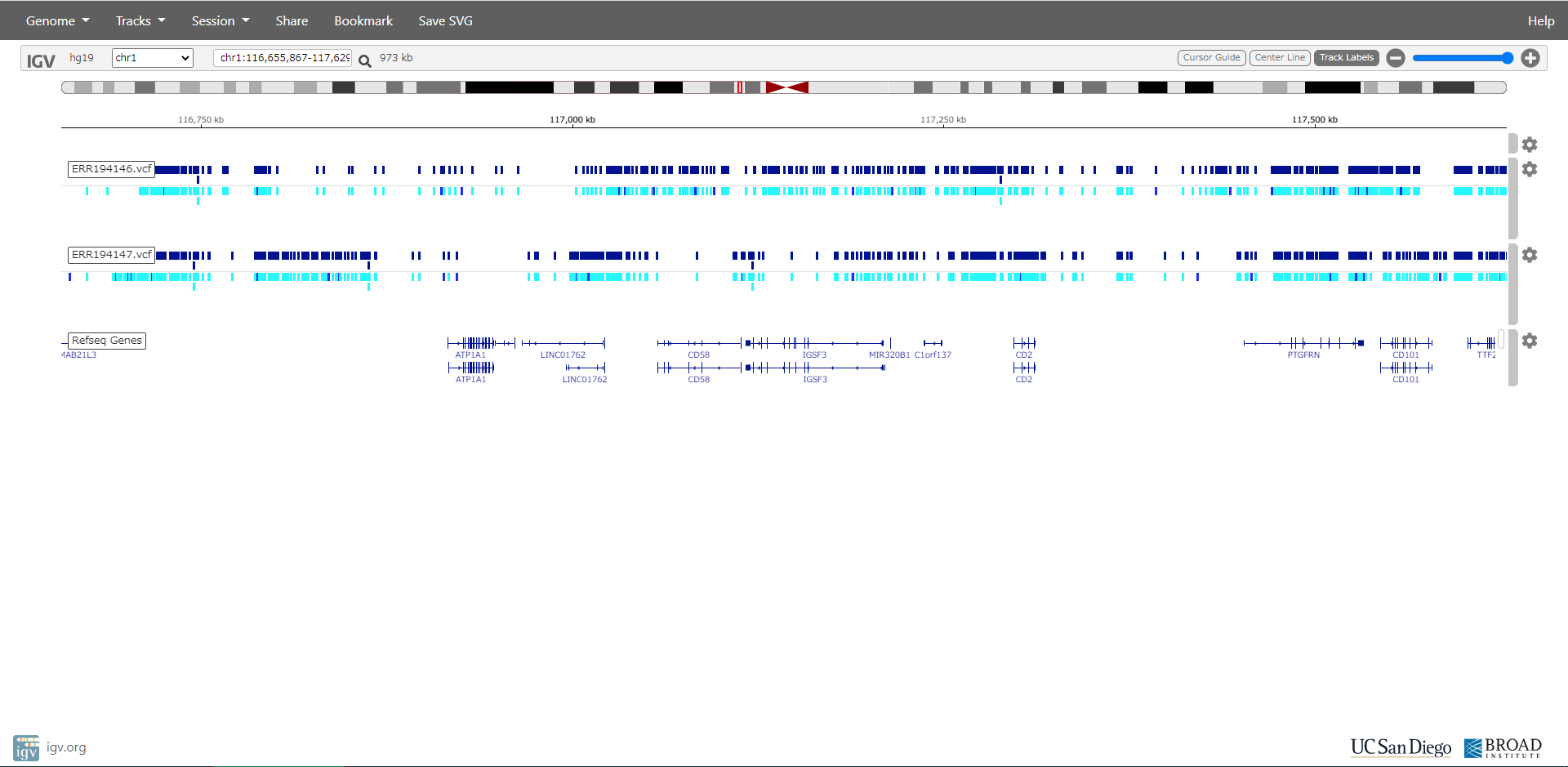
When all processes are done, ERR194146.vcf and ERR194147.vcf are created in ERR194146 and ERR194147 directory.
Sequences of primer pair are also outputted into vcf file.
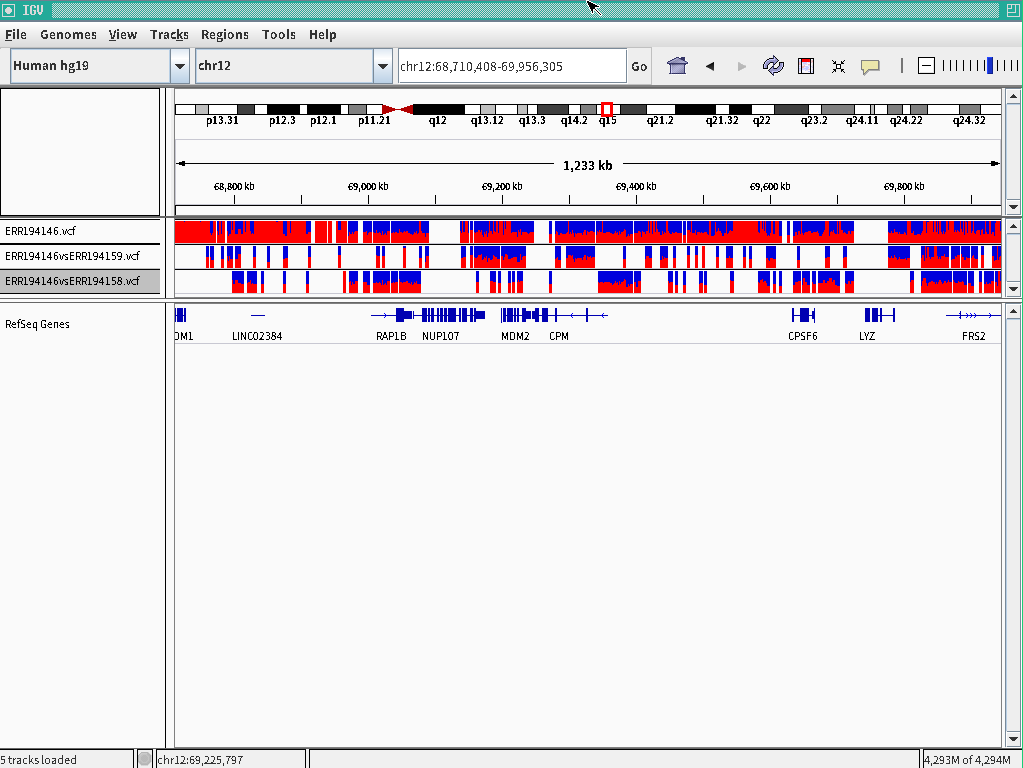
- If you have control sequences for the target,
$ perl ped.pl target=ERR194146,control=ERR194158,ref=hg19
$ perl ped.pl target=ERR194146,control=ERR194159,ref=hg19
Specific polymorphisms for the target will be outputted.

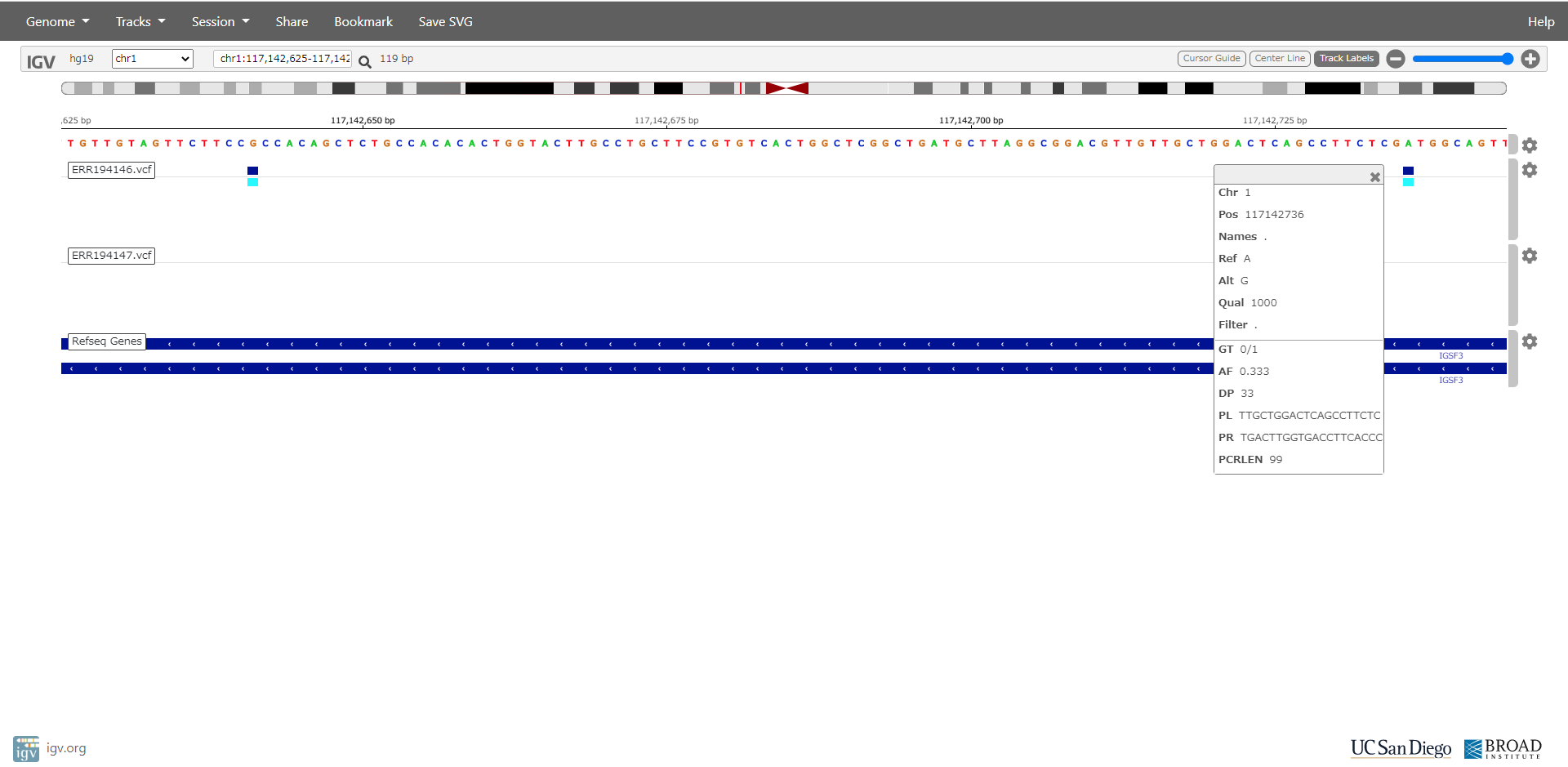
 Click to large.
Click to large.